Cancer functional genomics lab
Background
Recent technological advances include CRISPR based technologies as well as advances in screening technologies are revolutionising our ability to use functional genomics. Research in the Rosenbluh lab uses state of the art functional genomic tools including pooled CRISPR loss/gain of function screens and apply these technologies towards identifying genes associated with cancer and developing new approaches to treat cancer.

Advances in oligo synthesis technologies make it possible to synthesis up to 100,000 different sequences in just a few days. This enables very rapid generation of pooled sgRNA libraries that target every known human or mouse coding gene. Pooled libraries are then used to generate a pooled lentiviral library that is used to infect cells at low MOI (this insures that every infected cell receives only one virus). Following propagation genomic DNA is extracted and next generation sequencing is used to quantify sgRNA abundance.
Current project in the lab
SRP19 as a target in APC deleted cancers
APC is a tumour suppressor gene that is mutated in a large number of cancers. Most notably APC mutations are found in 80% of colorectal cancers. Despite the clear significance of APC in driving cancer currently no approaches are available to treat APC mutated cancers. Furthermore, studies in cultured cancer cell lines and animal models demonstrate that contentious suppression of APC is required for tumour growth even in advanced metastatic colon cancer. However, despite the clear evidence demonstrating the importance of APC in colon cancer pathogenesis we currently lack approaches for direct targeting of tumour suppressors. An alternative approach is to identify genes that are required for proliferation only in the context of APC loss of function mutation. As a proof of principle for this strategy, our previous work used genome scale shRNA loss of function proliferation screens and identified genes that are essential in cancers that harbour loss of a tumour suppressor gene. These experiments, identified a class of genetic vulnerabilities that are associated with DNA loss of a tumour suppressor we termed, CYCLOPS (Copy-number alterations Yielding Cancer Liabilities Owing to Partial losS). Specifically, heterozygous loss of a tumour suppressor is accompanied by heterozygous deletion of neighbouring cell essential genes and as a consequence, the mRNA and protein levels of these neighbouring cell essential genes is reduced. Suppressing the residual expression of the cell essential genes inhibits proliferation only in cells that harbour loss of the tumour suppressor gene. Using this strategy, we identified SRP19, a component of the signal recognition complex, as a CYCLOPS in cancers that harbour APC loss. Mechanistically, SRP19 is located in close physical proximity (15kb) to APC and is lost in cancers that harbour APC loss. Our preliminary data show that cell lines and tumours that harbour APC loss have lower levels of SRP19 mRNA and protein and are highly sensitive to additional suppression of SRP19 expression. These observations suggest inhibitors of SRP19 as a strategy against cancers with APC loss. The overall aim of this proposal is to evaluate SRP19 as a therapeutic target in a specific population of colon cancers that harbour APC loss (~20% of colon cancers) and to develop strategies to inhibit SRP19 as an approach to treat these cancers. More generally, although this project is focused on targeting of colon cancers with APC loss the same approach could in principle be applied for targeting any tumour suppressor that is lost in cancer.

Systematic identification and validation of breast cancer risk genes through follow up of genome-wide association studies.
Using genome wide association studies (GWAS) our collaborator Prof. Georgia Chenevix-Trench (QIMR, Brisbane) and her team have identified 179 breast cancer (BC) risk loci (Michailidou et al. Nature, 2017). The functional mechanism behind the associations usually involves perturbed regulation of target gene transcription by risk single nucleotide polymorphisms (SNPs) lying in regulatory elements positioned some distance from the target. The nearest gene to the GWAS ‘hit’ is not necessarily the target of the association, and for some loci there are multiple gene targets. This project uses large scale pooled clustered regularly interspaced short palindromic repeats (CRISPR) knockout and activation screens of all the predicted target coding and non-coding genes at these loci. We will evaluate the effect of over-expressing or suppressing the expression of all candidate BC risk genes on the ability of cells to proliferate in vitro, and in immune-deficient mice, as well as their ability to promote bypass of cellular senescence, in order to identify determinants of BC risk. In addition to enhancing our understanding of the genetic variants associated with BC risk, these experiments will enable new strategies for risk reduction therapies.

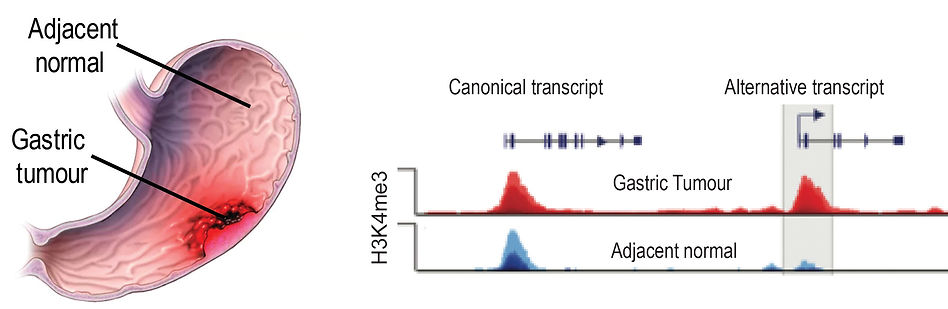
Identification of alternative transcripts that drive gastric cancer
Gastric cancer is a leading cause of mortality. Currently only a limited number gastric cancer treatments are available and new treatment options are necessary. Our collaborator Prof. Patrick Tan (NUS-Duke, Singapore) used ChIP-Seq from normal and gastric cancers and has identified a class of genes that in gastric cancers are expressed from an alternative promoter (Qamra et al Cancer Discovery, 2017). Using a verity of CRISPR based approaches we are identifying alternative transcripts that are essential for proliferation of gastric cancer and could be used as targets for development of gastric cancer specific therapies.
Heading 1
Identification and characterisation of functionally active circular RNAs
Circular RNAs (circRNAs) are a, recently discovered, covalently closed, single-stranded RNA molecules that are formed by mRNA back-splicing (Fig. 1A). circRNAs can be expressed at high levels (sometimes more than the linear form) and are tissue and disease specific, strongly suggesting that circRNAs have a functional role. However, apart from a very small number of cases, the functions of individual circRNAs are unknown. Using a previously described1 shRNA-based strategy we have developed a pooled-library targeting all known circRNAs (Fig. 1B). Our preliminary data, in three cell-lines, identify circRNAs that are essential for cell proliferation. To identify circRNAs that regulate core cell-essential processes or key signalling pathways we will use this library for a proliferation screen in a panel of 20 cell-lines that are dependent on constitutive activation of the WNT or MAPK pathways. By comparing circRNA dependencies amongst these cell-lines we will find pathway specific as well as core-essential circRNAs (Fig. 1C). This project will provide understanding into the functions of an abundant yet poorly understood molecule. Since these pathways play key roles in embryological growth and development, and are major sites of dysregulation in diseases, our findings will both enhance our understanding of developmental processes, and provide springboards for seeking new therapeutic approaches in a range of diseases.
